Current treatment methods for cardiac arrhythmia include medication, pacemakers, cardioversion, surgery, and
fluoroscopy-guided radiofrequency ablation. In particular, radiofrequency ablation is recommended in many cases,
due to its less invasive nature. However, there are still risks associated with it, such as injury to the heart,
including perforation through the muscle or damage to one of the valves within the heart, exposure to radiation,
which increases risk of cancer and genetic defects. MRI-guided radiofrequency ablation of the heart offers several
advantages over current techniques, such as elimination of radiation exposure.
Currently most of the visualization for such procedures is done by either 2-D fluoroscopy or manual segmentation
from 3-D CT models. On one hand, since fluoroscopy gives a 2-D projection of the 3-D object, it is difficult to accurately determine
a 3-D location. On the other hand, manual segmentation is time consuming and suffers from reliability and consistency
issues. Clearly, the improvement of 3-D visualization during the procedure could potentially help the physician
locate the target more efficiently as well as eliminate catheter-induced injury to the heart.
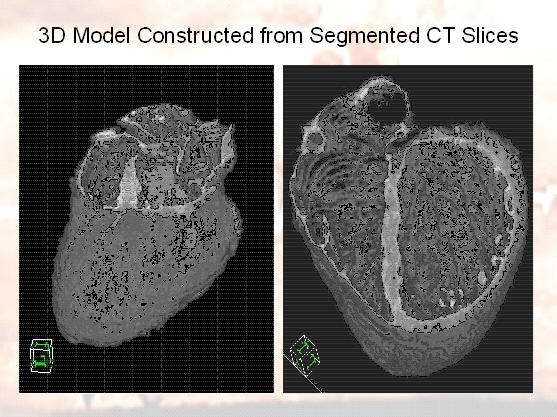
In this project, we are developing methods for automatically building and updating a 3-D model of the heart using real time
MR images. We first build a high-resolution 3-D model of the heart using MR and CT images acquired prior to the procedure.
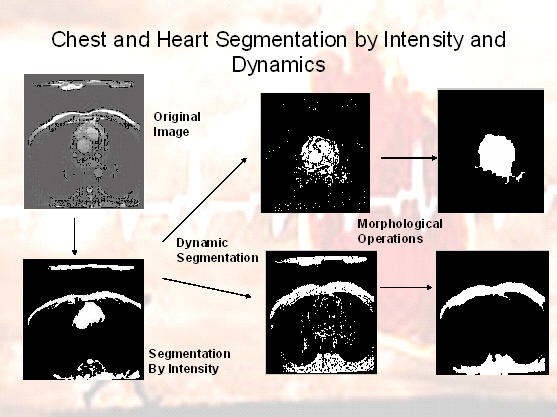
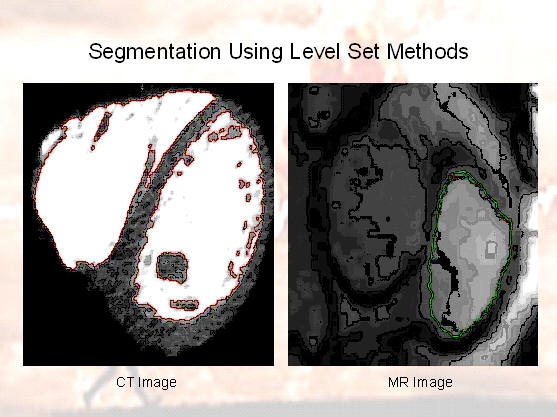
For accurate heart segmentation, two different methods have been presented: an fast algebraic method that does not impose
hard spatial coherence hence smoothness, and a level set method, which is iterative but imposing smoothness. Given MRI
data sequences, the heart and chest are separated from the background using the former method. In addition, we use
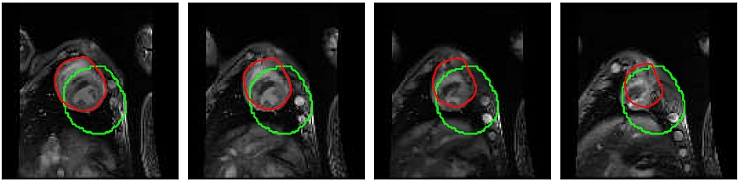
dymanic textures to segment regions with different motions. The latter is used to segment different regions from the heart as
well as to obtain smoother heart segmentation. We then build a 3-D high-resolution dynamic model that is updated with 2-D
low-resolution images acquired in real time. In brief, the problems we try to solve are:
- Algebraic Heart/Chest/Background Segmentation
- Level Set based Epicardial Segmentation
- Segmentation Within The Heart
- Building The 3-D Model