Project Summary
Nonlinear dimensionality reduction refers to the problem of finding
a low dimensional representation for a set of points lying on a
nonlinear manifold embedded in a high dimensional
space. This problem is fundamental to many problems in
computer vision, machine learning and pattern recognition, because
most datasets often have fewer degrees of freedom than the dimension
of the data. In computer vision, for example, the number of pixels
in an image can be rather large, yet most computer vision models use
only a few parameters to describe the geometry, photometry and
dynamics of the scene. The question of how to detect and represent
low dimensional structure in high dimensional data is fundamental to
many disciplines and several attempts had been made in the past in
different areas to address this.
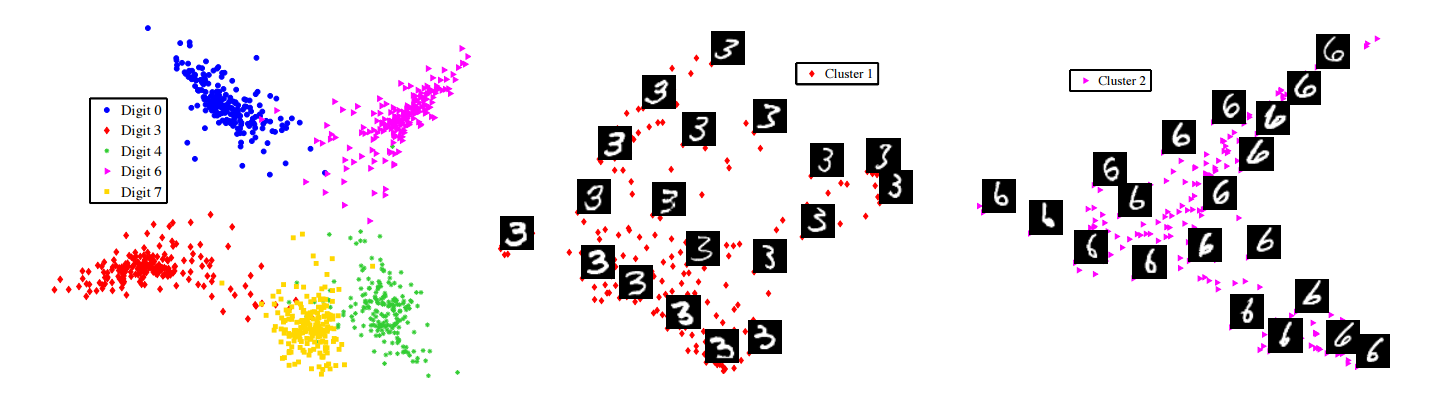
Over the past few years, various techniques have been developed for learning a low dimensional representation of a nonlinear manifold embedded in a high dimensional space. Although the goals of dimensionality reduction, classification and segmentation have always been intertwined with each other, considerably less work has been done on extending nonlinear dimensionality reduction techniques for the purposes of clustering data living on different manifolds. Unfortunately, most of these techniques are limited to the analysis of a single connected nonlinear manifold and suffer from degeneracies when applied to linear manifolds (subspaces). Figure 1 shows the types of manifolds to be segmentated into different groups.

Figure 1: Types of manifolds to be clustered.
Over the past few years, various techniques have been developed for learning a low dimensional representation of a nonlinear manifold embedded in a high dimensional space. Although the goals of dimensionality reduction, classification and segmentation have always been intertwined with each other, considerably less work has been done on extending nonlinear dimensionality reduction techniques for the purposes of clustering data living on different manifolds. Unfortunately, most of these techniques are limited to the analysis of a single connected nonlinear manifold and suffer from degeneracies when applied to linear manifolds (subspaces). Figure 1 shows the types of manifolds to be segmentated into different groups.

Figure 1: Types of manifolds to be clustered.